Document
1. General introduction
Rice is a major food for the world's population and serves as a model plant in cereal genome research. The Rice Information GateWay (RIGW) was constructed in 2016 upon the completion of the Oryza sativa ssp. indica cv. Zhenshan 97 and Minghui 63 genomes sequenced by Huazhong Agricultural University (HZAU) and Arizona Genomics Institute (AGI) with the aim of providing accurate and timely annotation of indica rice genome sequences. We have annotated the indica rice genomes ZS97 and MH63 for gene content, repetitive elements, non-coding RNA, SNPs, inversions, translocations, presence/absence variations, and segmental duplications, etc. Sequence polymorphisms between different rice subspecies indica and japonica have been identified. Designed as a basic platform for indica rice study, RIGW presents the sequenced genomes and related information in systematic and graphical ways for the convenience of in-depth comparative studies.
Table 1. Summary of genomic features in RIGW
| ZS97RS1 | MH63RS1 | |
|---|---|---|
| Total size of assembled contigs (bp) | 346,854,256 | 359,918,891 |
| Number of contigs | 237 | 181 |
| Contig N50 (bp) | 2,339,070 | 3,097,358 |
| GC content | 43.59% | 43.63% |
| Numbers of gene models/transcripts | 54,831/78,033 | 57,174/80,581 |
| Number of non-TE gene loci | 34,610 | 37,324 |
| Mean transcript length (bp) | 1,955 | 1,962 |
| Mean coding sequence length (bp) | 1,348 | 1,368 |
| Number of predicted PPI | 1,296,100 | 1,155,115 |
| Number of homologous genic blocks | 241 | 238 |
| Number of KEGG mapped transcripts | 11,272 | 11,334 |
| Number of ortholog genes | 29,814 | 30,980 |
2. Homepage
In the homepage, Quick Link can help to quickly connect to the commonly used function modules, MapView can display the location distribution of users.

3. Gene Search
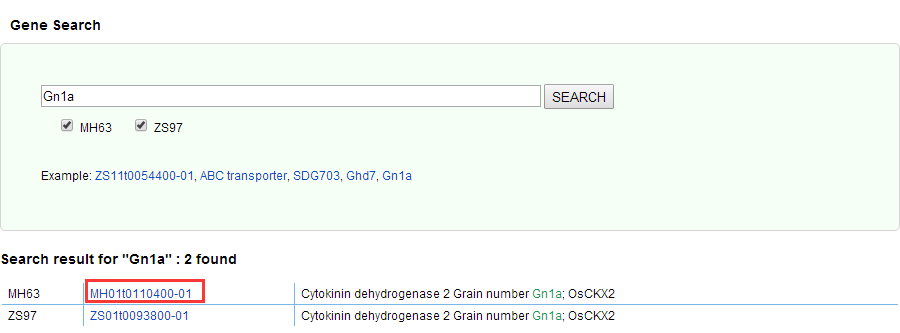
Click on "Gene locus", you can search your desired functional gene. You can type in any one of the fields such as gene function, gene name and gene Locus ID, such as "ABC transporter", "Gn1a" and "ZS11t0054400-01".

Gene details page needs to be found in the results of Gene Search:
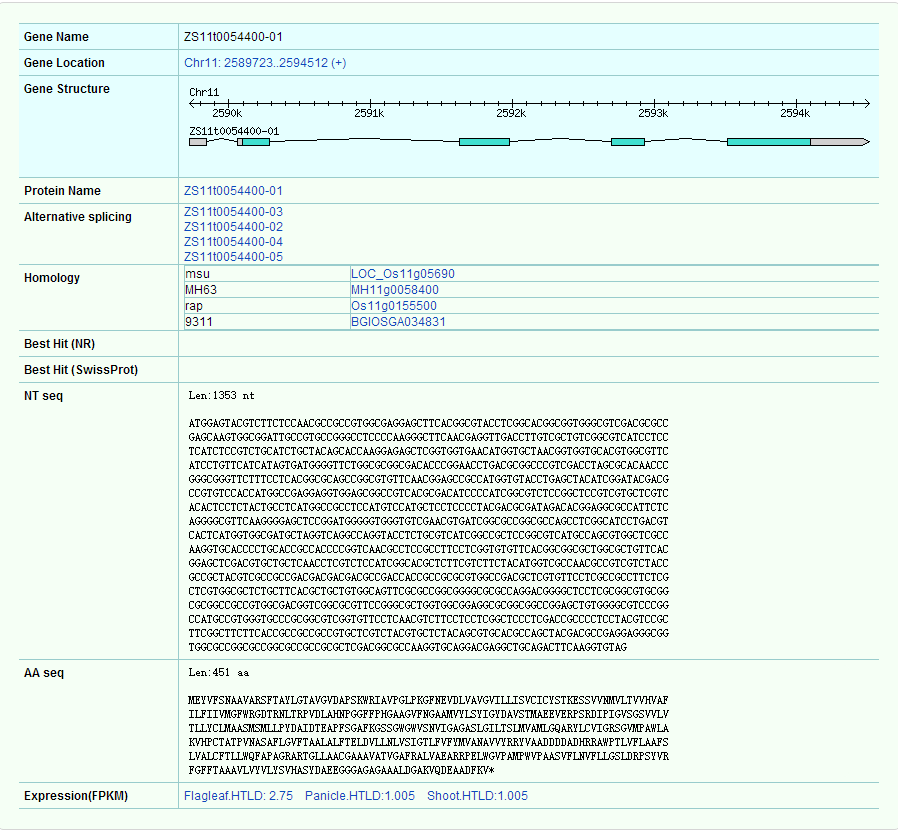
The gene details page shows the location of the gene first (can be linked to Gbrowse by hyperlink), gene structure, variable shear form, homologous gene, nucleic acid sequence, protein sequence and expression information (You will see more information in the Expression module via hyperlink).
As well as the GO annotation and domain analysis of the gene, click on the corresponding category ID to link to the external database.

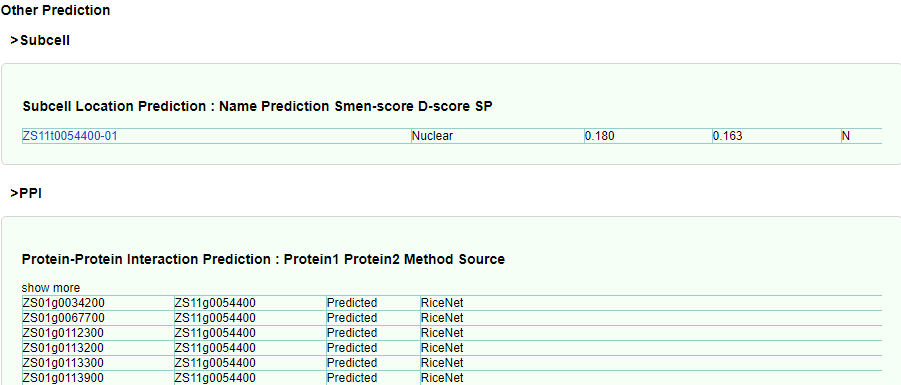
Finally, subcellular localization analysis and PPI prediction were added, and the database will show more annotation information related to gene function with update.
4. BLAST
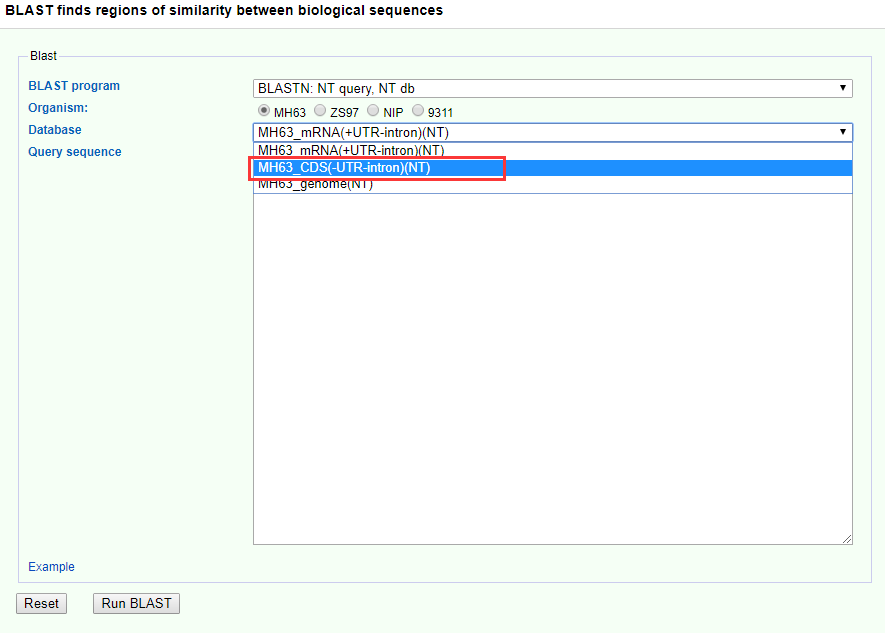
BLAST as a fast basic local alignment search tool, in the functional genome database is indispensable. You can click on Search-> BLAST to enter the module, the current comparison of the reference species include MH63, ZS97, NIP, 9311. If you think there are other very meaningful and commonly used rice genomes should be added, please send email to us. It is worth noting that, based on the sequence level you are looking for, you should choose the correct alignment sequence library, for example, the query sequence is CDS sequence, then the database is best to use CDS (-UTR-intron).
5. Sequence Fetch
On the Sequence Fetch page, the corresponding sequence information can be obtained from the gene Locus or the genome coordinates. For the gene sequence, select the pre-mRNA, mRNA, protein to play the corresponding sequence.

6. Clone Gene
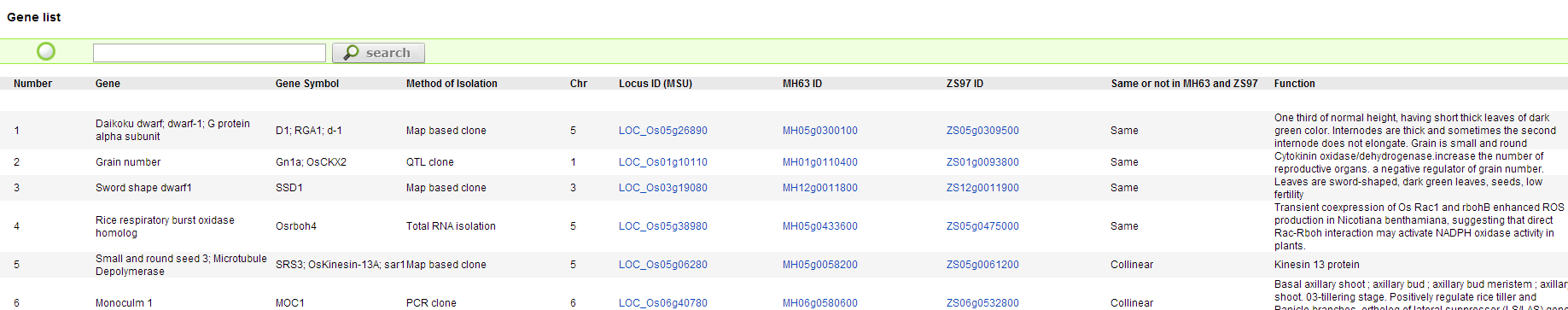
Clone Gene page can query the information of cloned genes in rice and its differences in two indica rice subspecies by Gene Locus, Gene Name and gene function keywords.
7. Metabolite
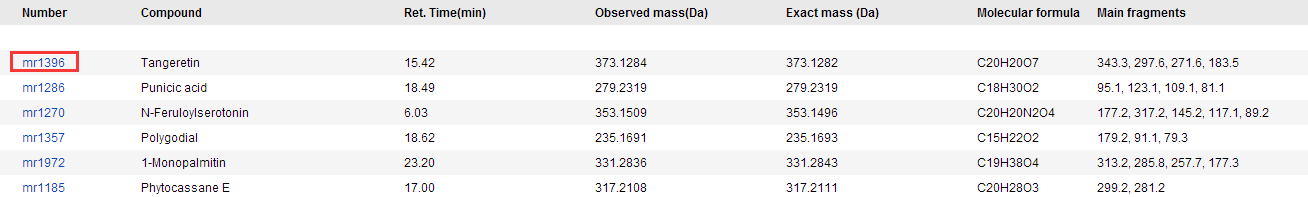
The Metabolite page describes the metabolites information that has been identified in rice and can be searched by compound name and metabolite ID. You can view more detailed information on the compound in ChEBI by clicking on the hyperlink.
8. Gene Expression
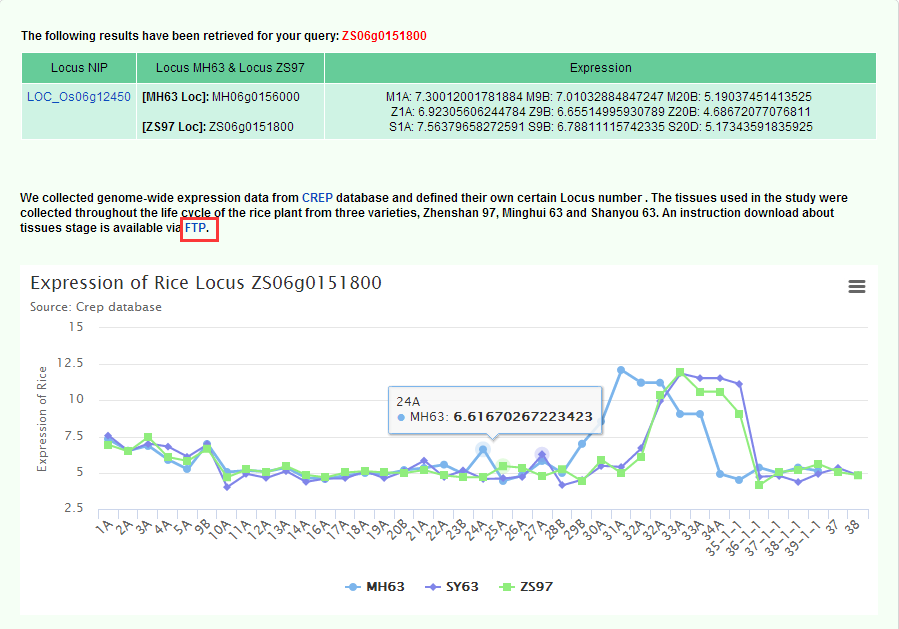
On Gene Expression page, We collected genome-wide expression data from CREP database and defined their own certain Locus number . The tissues used in the study were collected throughout the life cycle of the rice plant from three varieties, Zhenshan 97, Minghui 63 and Shanyou 63. You can query the corresponding sample information via the Locus number of the MH63, ZS97, or NIP and download the introductory information.
9. PPI
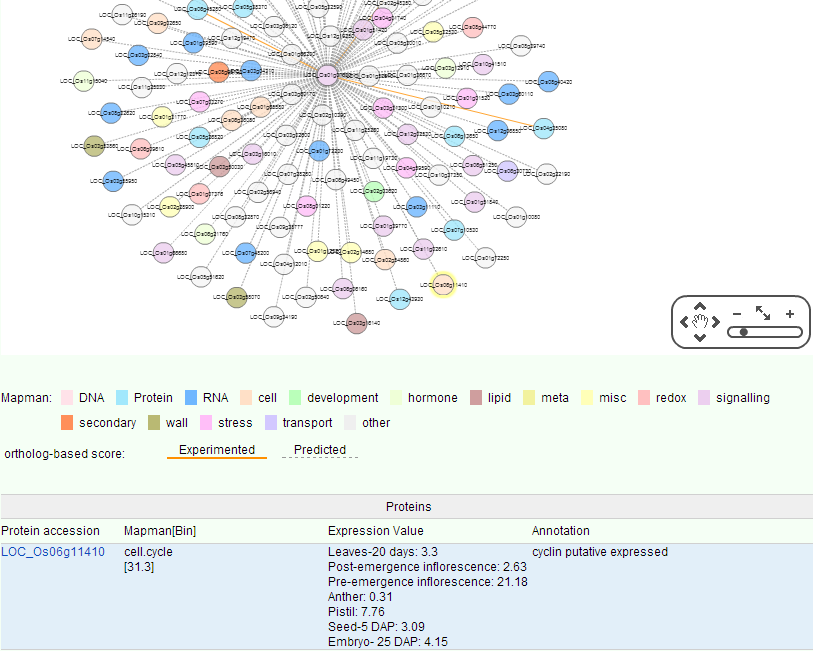
Proteins seldom perform their biological functions independently, and most complex cellular processes must be understood via large-scale PPI network. Genome-wide PPI networks have become powerful tools to study the cellular behaviors with a global view, and they can reveal the relationships between different kinds of proteins with various functions. In total, 1871563 rice PPIs were collected from database as PRIN, RiceNet, IntAct and some related articles, among them 929 PPIs were experimented with yeast two-hybrid system. In PPI-tools, we visualize these interactions with different color nodes to represent the different pathway classifications of the proteins. Related data will be continuously updated. In the query box you can also type two Locus to observe the relationship between its PPI network. Click on the points and lines in the network to display more details below.
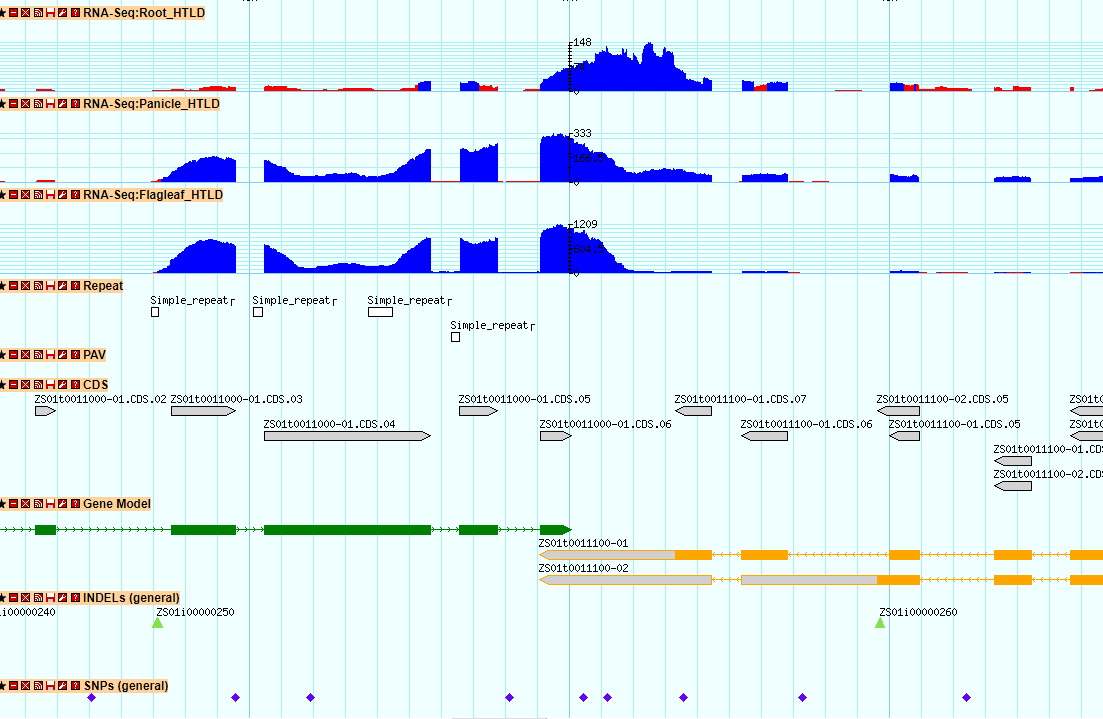
10. GBrowse AND Gbrowse_syn
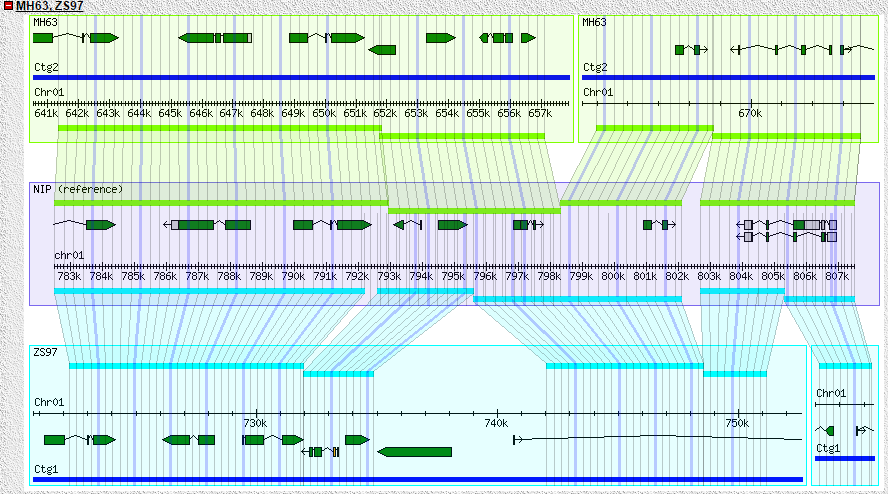
GBrowse is a powerful genomic data visualization tool. We run it in the MH63, ZS97 genomic structure display, you can by selecting a different track see the specific region of the different genomic content. In order to show the difference between the two subspecies, we also added information such as PVs, SNPs and InDels. You can click the feature, the corresponding region can also be showed in another gene. The Gbrowse_syn tool is used to show collinear regions and genes between two genomes. For more detailed use of GBrowse you can refer to http://gmod.org/wiki/GBrowse.
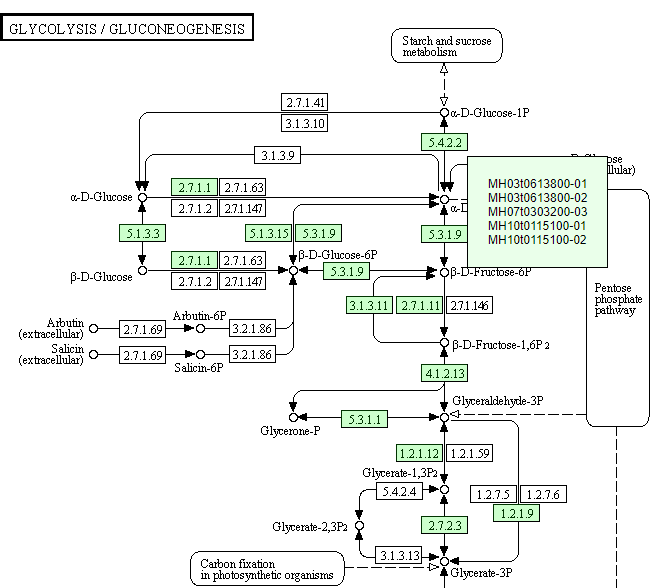
11. Pathway
In the Pathway module, we map the gene to the metabolic pathway and show it based on the KEGG Map.
12. Homologous Region
We defined homologous gene pairs with MCscanX ( e-value: 1e-10) between chromosomes, and determined the homologous genic blocks based on the distance to show the traces of the original rice genome duplication in evolution. In addition, we have drawn the link map of homologous genic blocks and attached hyperlinks to gbrowser in the related area.

13. CRISPR design
The CRISPR/Cas9 technology is a modern, fashionable method in plant research. CRISPR design is based on CRISPR-P 2.0 tool, and it can design sgRNA for almost all available rice genomes. The parameters described with reference to http://crispr.hzau.edu.cn/CRISPR2/help.php.
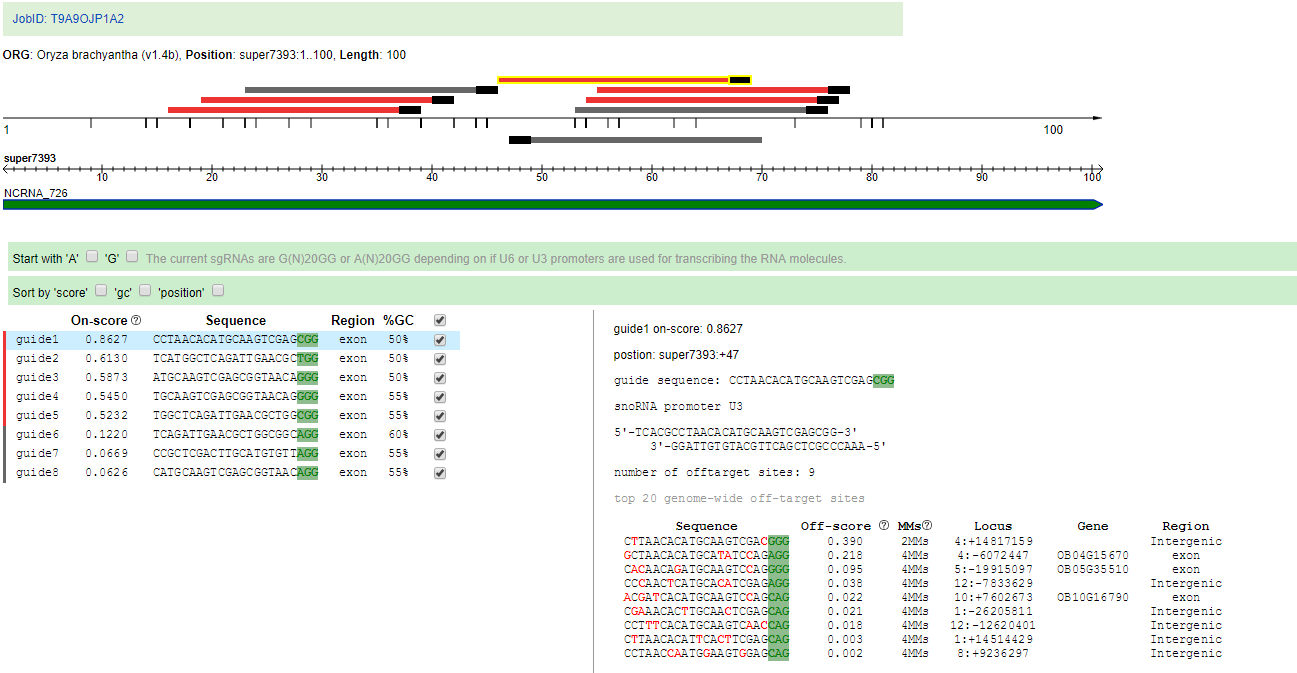
14. Orthologous
Orthologous Groups among Oryza indica, Oryza japonica, Arabidopsis, Brachypodium, Maize, Poplar and Grapevine. You can do orthologous groups searches using for different species locus (MH07g0048400/ZS07g0055600/LOC_Os07g05400) by submitting your query in the search box above.
15. KEGG/GO Enrichment
You can annotate KEGG/ GO of your Locus list on KEGG/ GO enrichment page. Furthermore, it is based on the genome-wide background, and can calculate the function enrichment of your provided sub-gene set (transcript id) .
16. ID Conversion
Based on the information of the whole genome pairing, the homology blocks between the two genomes were identified. The optimal mapping results were selected as the candidate mapping gene for the gene in another species by selecting the longest protein sequence within the homologous block for blastp (evlaue <10-5, identify> 80%). For the candidate mapping gene pairs in four species, mutual evaluation and manual examination will be used in correction, and finally we identified gene corresponding list among ZS, MH, NIP and 9311. The correspondence between MSU and RAP locus was obtained from RAP-DB.
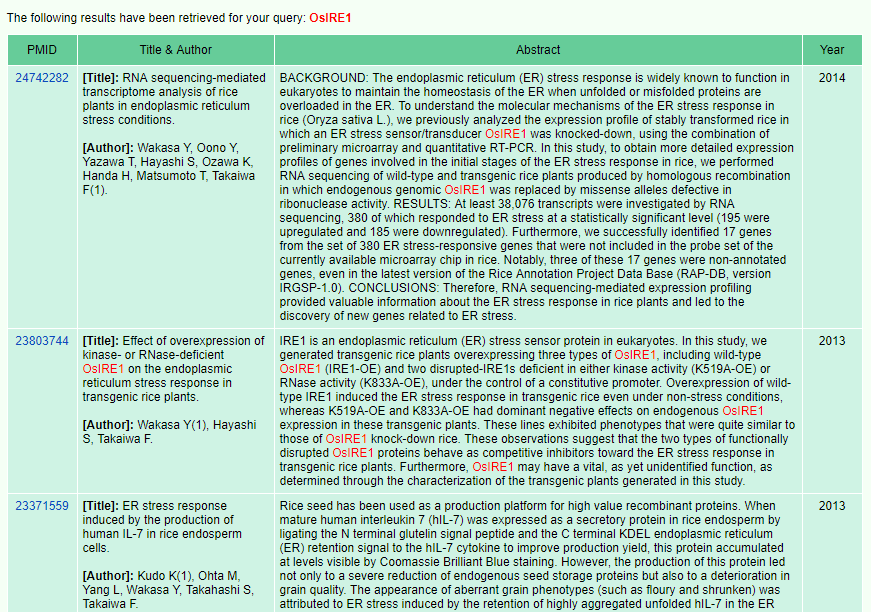
17. Literature
Lastly, a text mining tool was available in RIGW, which allows a user to search papers by gene names or keywords in 27,831 rice-related literatures obtained from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/).
18. Contact us
If you have questions, please contact Ling-Ling Chen (llchen@mail.hzau.edu.cn) or Jianwei Zhang (jzhang@mail.hzau.edu.cn, jzhang@cals.arizona.edu).
